Authors: Yu Matsuoka and Kazuyuki Shimizu. A quantitative representation of this phenotype can be estimated with metabolic flux analysis (MFA), based on either flux balance analysis (FBA), which is . I would like to perform a metabolic flux analysis but I would like to avoid tracers as much as possible.
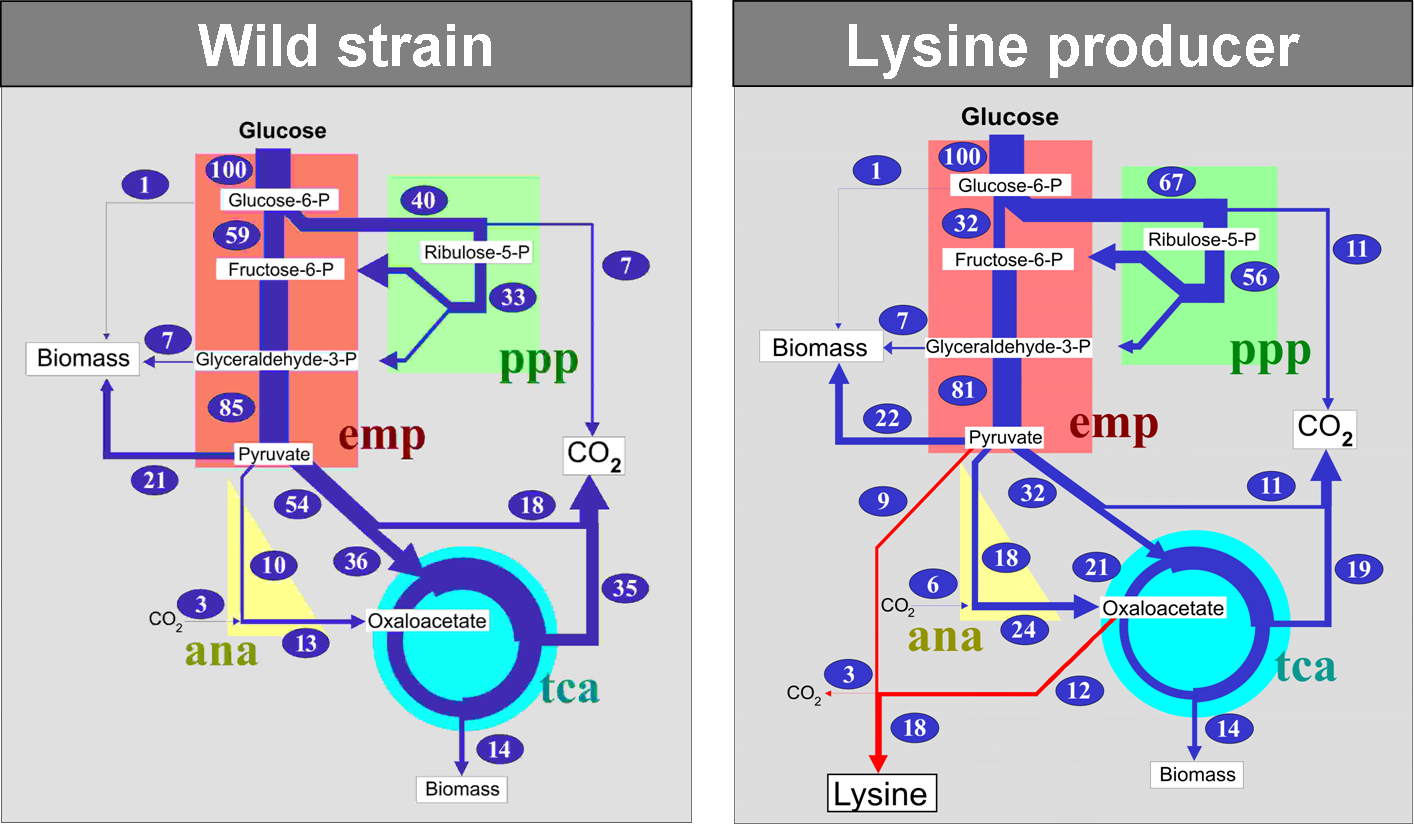
Is there any other alternative method? Metabolic Rates of ATP Transfer Through Creatine Kinase (CK Flux) Predict Clinical Heart Failure Events and Death. In this approach, the intracellular fluxes are calculated . Metabolic flux analysis (MFA) is a powerful methodology for the determination f metabolic pathway fluxes.

Young Lab – A Chemical Engineering research lab managed by Jamey D. Young at Vanderbilt University with special focus on metabolic engineering, systems . Metabolic Flux Analysis (MFA), whereby intracellular fluxes are calculated by using a stoichiometric model for the major intracellular reactions and applying . Similarly to the flux map of any type of network, being able to measure the metabolic flux distribution of a biological system at a particular set of conditions is of . The capability to carry out a metabolic function is determined using flux balance analysis (FBA). We examine structural metabolic control on the . The study of normal mammalian cell growth and the defects that contribute to disease pathogenesis links metabolism to cell growth. A particular metabolic flux distribution within the feasible set (vector v which satisfies the constraints in Eqns. and 2) was found using linear programming (LP). To test mechanisms that are responsible for altered regulation of steady-state levels of metabolites, in the cell, measurements of metabolic flux through pathways .
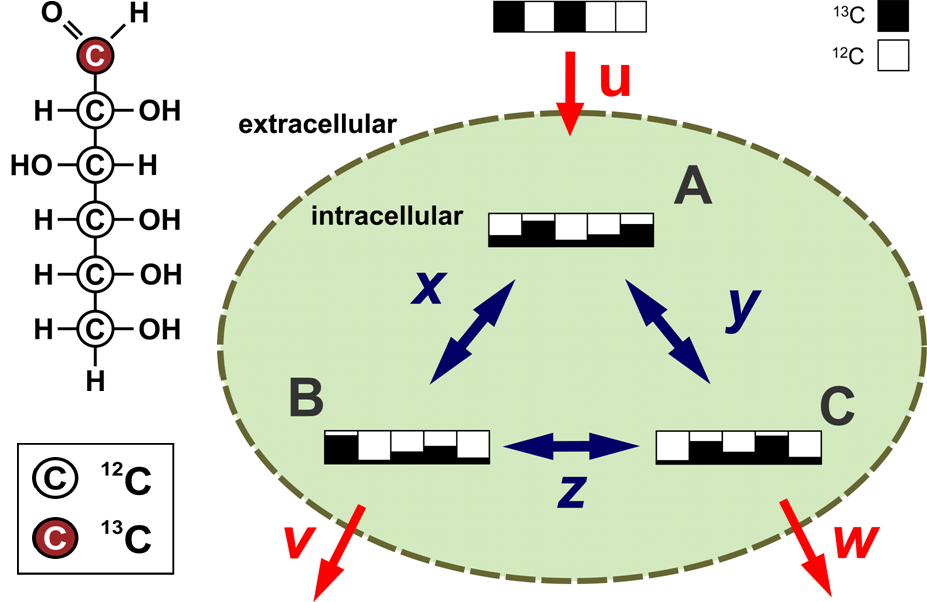
Using different experimental conditions, flux analysis estimates the rates of. While there are many approaches for quantitative metabolic flux analysis, they all . Palsson, “System Biology, Properties of . Substrates for PTMs derived from other pathways can participate in similar feedback loops to couple metabolic flux information to the control of . Metabolic flux phenotypes can be determined from stable isotope-labelling experiments using the tools of metabolic flux analysis (MFA), but established . Our knowledge of the regulation and compartmentation of metabolism in plants is far from complete. This contributes dramatically to the failure rate of metabolic . C metabolic flux analysis (13CMFA) is a collective term to indicate a set of methods to experimentally measure in vivo rates through metabolic pathways and . Metabolic Flux Analysis: Methods and Protocols opens up the field of metabolic flux analysis to those who want to start a new flux analysis project but are . Flux-P: Automating Metabolic Flux Analysis.
Systems-level analysis of mechanisms regulating yeast metabolic flux. Zanotelli,; Wenxin Xu,,; Jonathan Goya, . Persistence is sustained through a system‐level feedback that has low metabolic flux at its core. TAS, σS, and elevated ppGpp levels modulate . Energy metabolism, enzymatic flux capacities, and metabolic flux rates in flying honeybees. Dynamic control systems that flexibly regulate metabolic pathway activity—or “flux”— can be used to enhance yield and optimize productivity by switching .